In constricted regions e. More diffusion yields a darker pixel because you lose signal as the water molecule can go anywhere. Once all the registrations have finished running, you are ready to move onto the next step. A thresholded skeleton mask image and derived distance map are then derived from this. The script finishes by telling you to check whether a suitable threshold for the mean FA skeleton is 0. 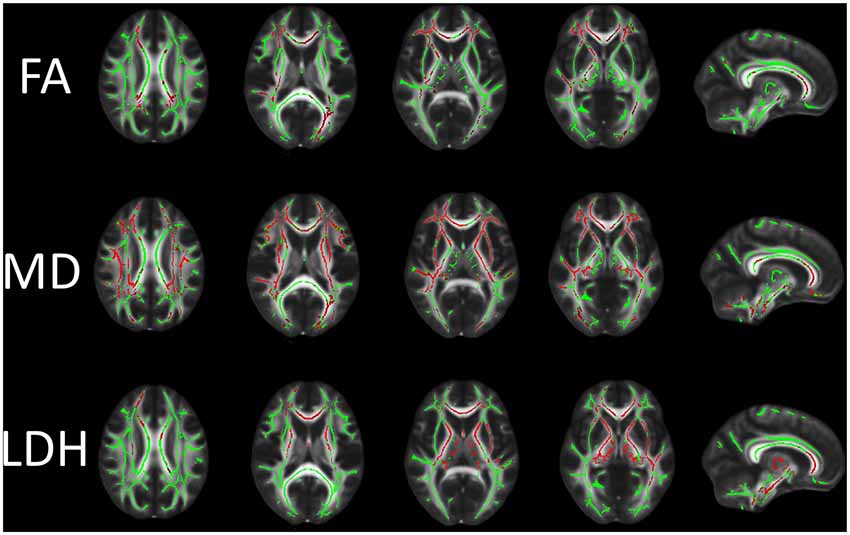
| Uploader: | Zulkishakar |
| Date Added: | 11 July 2012 |
| File Size: | 35.41 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 55794 |
| Price: | Free* [*Free Regsitration Required] |
On the other hand, in some areas such as along the fiber bundles of neurons e. Second, this point can then be "inverse warped" back into the subject's native space, by inverting the nonlinear registration that was originally applied. For example, if you are using bedpostX outputs, copy the dyads dyads1,dyads2,etc.
Sample DTI processing using FSL
Before running randomise you will need to generate a design matrix file, e. For your convenience, we provide example text short and more tbsss versionswhich you are welcome to use in your methods description. You can vary the amount of diffusion weighting level of sensitivity to diffusion. Alternatively you can use the Glm GUI to generate these design matrix and contrast files. To complete this tutorial you will want the following: There are 3 eigenvectors in each direction: The second is to take a skeleton-space voxel or set of voxels for example, a skeleton-space blob that is significantly different between the two groups of subjects tbzs the study back to the space of the original subjects' data, in order to run tractography for each subject, with that result forming the tractography seed mask.
You probably want to change the colourmap, for example to Green, and increase the transparency with the transparency slider so that when you load the stats image in, it is easier to see.
Each subject's aligned FA data was then projected onto this skeleton and the resulting data fed into voxelwise cross-subject statistics. Z-statistic or p-values you should not use the -n option.

ybss This now becomes the recommended target for the nonlinear registrations, instead of registering every subject to every other subject, and choosing the most typical. If your data has continuous values e. The following instructions assume that you are using bedpostX estimates of fibres orientations dyads1,dyads2,etc. TBSS aims to solve these issues via a carefully tuned nonlinear registration, followed by b projection onto an alignment-invariant tract representation the "mean FA skeleton".
More diffusion yields a darker pixel because you lose signal as the water molecule can go anywhere.
Diffusion Tensor Imaging Analysis using FSL
The script then takes the target and affine-aligns it into 1x1x1mm MNI space - this resolution is chosen as the later skeletonisation and projection steps work well at this resolution, and the choice of working in MNI space is chosen for convenience of display and coordinate reporting later. This option is applied by using the -T flag. If the diffusion value is low, it is because the molecules are blocked csl something else.
In comparison, the all-subjects-to-all-subjects option takes about 5 minutes x N x N.
The upper level should probably be set to something like 0. To run such analysis, first complete the above TBSS processing steps First, the original asymmetric skeleton is dilated thickened by one voxel, and saved to a temporary file.
There are multiple volumes, but each at a different diffusion direction. This is then skeletonised to generate the initial symmetric skeleton. It also saves the three orthogonal vectors V1, V2, V3.
The nonlinear warps and skeleton projection can then also be applied to other images such as the second eigenvalue.
A tensor is arranged as a matrix. In DTI, you use a tensor tbbss than a vector. Higher ADC values mean it can go very far without interruption, and it takes the signal with it. For example, you may use partial volume fraction estimates from bedpostX instead of FA at each voxel.
If all the processing so far has worked ok the skeleton should look like the examples shown here see the TBSS paper for more examples of different subjects' results underneath the skeleton.
But in tractography, you would say the more the merrier to get the better image. The other is to use hbss data for tractography to illustrate the shape and direction of fiber tracts comparing C versus D.

Комментарии
Отправить комментарий